5.11. pyopus.problems.cutermgr — CUTEr problem manager¶
CUTEr problem manager
Currently works only under Linux.
CUTEr is a collection of problems and a systef for interfacing to these problems. Every problem is described in a .sif file. This file is converted to FORTRAN code (evaluators) that evaluate the function’s terms and their derivatives. CUTEr provides fortran subroutines (CUTEr tools) that call evaluators and return things like the function’s value or the value of the function’s gradient at a given point, etc. CUTEr tools are available in a static library libcuter.a.
Every CUTEr problem is first compiled with a tool named sifdecode that produces the evaluators in FORTRAN. The evaluators are compiled and linked with the CUTEr tools and the Python interface to produce a loadable binary module for Python. The evaluators must be compiled with the same compiler as CUTEr tools. The loadable Python module is stored in a cache so that it does not have to be recompiled every time one wants to use the CUTEr problem.
Installing CUTEr
Before you use this module, you must build sifdecode and the CUTEr tools library.
Unpack the SIF decoder, build it by running install_sifdec (answers to the first five questions should be: PC, Linux, gfortran, double precision, large). Set the SIFDEC enavironmental variable so that it contains teh path to the folder where you unpacked the sifdecode source (where install_sifdec is located). After sifdecode is built a subfolder named SifDec.large.pc.lnx.gfo is created in $SIFDEC. Set the MYSIFDEC environmental variable so that it contains the path to this folder. Add $MYSIFDEC/bin to PATH so sifdecode will be accessible from anywhere. Also add $SIFDEC/common/man to the MANPATH environmental variable.
Unpack CUTEr. The tools library (libcuter.a) must be built with -fPIC (as position independent code). To achieve this, the file config/linux.cf in CUTEr installation must be edited and the -fPIC option added to lines that begin with:
#define FortranFlags ...
#define CFlags ...
Build the CUTEr library by running install_cuter (answers to the first five questions should be: PC, Linux, gfortran, GNU gcc, double precision, large). Set the CUTER environmental variable so that it contains the path to the folder where you unpacked CUTEr (the one where install_cuter is located). After CUTEr is compiled (i.e. libcuter.a is built) a subfolder named CUTEr.large.pc.lnx.gfo is created. Set the MYCUTER environmental variable so that it contains the path to this folder. Add $CUTER/common/man to MANPATH $CUTER/common/include to C_INCLUDE_PATH, $MYCUTER/double/lib to LIBPATH, and $MYCUTER/bin to PATH.
On 64-bit systems use the standard version of CUTEr (not the 64-bit one).
Download SIF files describing the available problems from http://www.cuter.rl.ac.uk/ and create a SIF repository (create a directory, unpack the problems, and set the MASTSIF environmental variable to point to the directory holding the SIF files).
The Python interface to CUTEr
Create a folder for the CUTEr cache. Set the PYCUTER_CACHE environmental
variable to contain the path to the cache folder. Add $PYCUTER_CACHE to the
PYTHONPATH envorinmental variable. The last step will make sure you can
access the cached problems from Python.
If PYCUTER_CACHE is not set the current directory (.) is used for caching.
Problems are cached in $PYCUTER_CACHE/pycuter.
The interface depends on gfortran and lapack. Make sure they are installed.
Once a problem is built only the following files in
$PYCUTER_CACHE/pycuter/PROBLEM_NAME are needed for the problem to work:
_pycuteritf.so– CUTEr problem interface module__init__.py– module initialization and wrappersOUTSDIF.d– problem information
A cached problem can be imported with from pycuter import PROBLEM_NAME. One
can also use importProblem() which returns a reference to the problem module.
Available functions
clearCache()– remove Python interface to problem from cacheprepareProblem()– decode problem and build Python interfaceimportProblem()– import problem interface module from cache (prepareProblem must be called first)isCached()– returnsTrueif a problem is in cache
CUTEr problem structure
CUTEr provides constrained and unconstrained test problems. The objective function is a map from Rn to R:
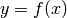
where 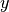 is a real scalar (member of
is a real scalar (member of 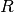 ) and
) and 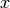 is a
n-dimensional real vector (member of
is a
n-dimensional real vector (member of 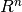 ). The i-th component of
). The i-th component of
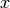 is denoted by
is denoted by 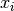 and is also referred to as the i-th
problem variable. Problem variables can be continuous (take any value from
and is also referred to as the i-th
problem variable. Problem variables can be continuous (take any value from
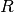 ), integer (take only integer values) or boolean (only 0 and 1
are allowed as variable’s value).
), integer (take only integer values) or boolean (only 0 and 1
are allowed as variable’s value).
The optimization problem can be subject to simple bounds of the form
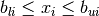
where 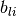 and
and 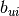 denote the lower and the upper bound
on the i-th component of vector x. If there is no lower (or upper) bound on
denote the lower and the upper bound
on the i-th component of vector x. If there is no lower (or upper) bound on
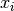 CUTEr reports
CUTEr reports 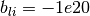 (or
(or 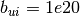 ).
).
Beside simple bounds on problem variables some CUTEr problems also have more sophisticated constraints of the form
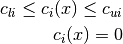
where 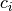 is the i-th constraint function. The former is an inequality
constraint while the latter is an equality constraint. CUTEr problems have
generally m such constraints. In
is the i-th constraint function. The former is an inequality
constraint while the latter is an equality constraint. CUTEr problems have
generally m such constraints. In 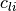 and
and 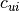 the values
-1e20 and 1e20 stand for
the values
-1e20 and 1e20 stand for 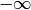 and
and 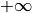 . For equality
constraints
. For equality
constraints 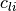 and
and 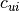 are both equal to 0. All
constraint functions
are both equal to 0. All
constraint functions 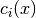 are joined in a single vector-valued
function (map from
are joined in a single vector-valued
function (map from 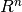 to
to 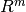 ) named
) named 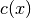 .
.
CUTEr can order the constraints in such manner that equality constraints appear before inequality constraints. It is also possible to place linear constraints before nonlinear constraints. This of course reorders the components of c(x). Similarly variables (components of x) can also be reordered in such manner that nonlinear variables appear before linear ones.
The Lagrangian function, the Jacobian matrix, and the Hessian matrix
The Lagrangian function is defined as
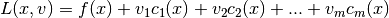
Vector v is m-dimensional. Its components are the Lagrange multipliers.
The Jacobian matrix (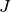 ) is the matrix of constraint gradients.
One row corresponds to one constraint function
) is the matrix of constraint gradients.
One row corresponds to one constraint function 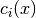 . The matrix
has n columns. The element in the i-th row and j-th column is the derivative
of the i-th constraint function with respect to the j-th problem variable.
. The matrix
has n columns. The element in the i-th row and j-th column is the derivative
of the i-th constraint function with respect to the j-th problem variable.
The Hessian matrix (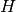 ) is the matrix of second derivatives. The
element in i-th row and j-th column corresponds to the second derivative with
respect to the i-th and j-th problem variable. The Hessian is a symmetric
matrix so it is sufficient to know its diagonal and its upper triangle.
) is the matrix of second derivatives. The
element in i-th row and j-th column corresponds to the second derivative with
respect to the i-th and j-th problem variable. The Hessian is a symmetric
matrix so it is sufficient to know its diagonal and its upper triangle.
Beside Hessian of the objective CUTEr can also calculate the Hessian of the Lagrangian and the Hessians of the constraint functions.
The gradient (of objective, Lagrangian, or constraint functions) is always taken with respect to the problem’s variables. Therefore it always has n components.
What does the CUTEr interface of a problem offer
All compiled test problems are stored in a cache. The location of the cache
can be set by defining the PYCUTER_CACHE environmental variable. If no
cache location is defined the current working directory is used for caching
the compiled test problems. The CUTEr interface to Python has a manager
module named cutermgr.
The manager module (cutermgr) offers the following functions:
clearCache()– remove a compiled problem from cacheprepareProblem()– compile a problem, place it in the cache, and return a reference to the imported problem moduleimportProblem()– import an already compiled problem from cache and return a reference to its module
Every problem module has several functions that access the corresponding problem’s CUTEr tools:
getinfo()– get problem descriptionvarnames()– get names of problem’s variablesconnames()– get names of problem’s constraintsobjcons()– objective and constraintsobj()– objective and objective gradientcons()– constraints and constraints gradients/Jacobianlagjac()– gradient of objective/Lagrangian and constraints Jacobianjprod()– product of constraints Jacobian with a vectorhess()– Hessian of objective/Lagrangianihess()– Hessian of objective/constrainthprod()– product of Hessian of objective/Lagrangian with a vectorgradhess()– gradient and Hessian of objective (if m=0) or gradient of objective/Lagrangian, Jacobian, and Hessian of Lagrangian (if m > 0)scons()– constraints and sparse Jacobian of constraintsslagjac()– gradient of objective/Lagrangian and sparse Jacobiansphess()– sparse Hessian of objective/Lagrangianisphess()– sparse Hessian of objective/constraintgradsphess()– gradient and sparse Hessian of objective (if m=0) or gradient of objective/Lagrangian, sparse Jacobian, and sparse Hessian of Lagrangian (if m > 0)report()– get usage statistics
All sparse matrices are returned as scipy.sparse.coo_matrix objects.
How to use cutermgr
First you have to import the cutermgr module:
from pyopus.problems import cutermgr
If you want to remove the HS71 problem from the cache, type:
cutermgr.clearCache('HS71')
To prepare the ROSENBR problem, type:
cutermgr.prepareProblem('ROSENBR')
This removes the existing ROSENBR entry from the cache before rebuilding the
problem interface. The compiled problem is stored as ROSENBR in cache.
Importing a prepared problem can be done with cutermgr:
rb=cutermgr.importProblem('ROSENBR')
Now you can use the problem. Let’s get the information about the imported problem and extract the initial point:
info=rb.getinfo()
x0=info['x']
To evaluate the objective function’s value at the extracted initial point, type:
f=rb.obj(x0)
print "f(x0)=", f
To get help on all interface functions of the previously imported problem, type:
help(rb)
You can also get help on individual functions of a problem interface:
help(rb.obj)
The cutermgr module has also builtin help:
help(cutermgr)
help(cutermgr.importProblem)
Storing compiled problems in cache under arbitrary names
A problem can be stored in cache using a different name than the original
CUTEr problem name (the name of the corresponding SIF file) by specifying the
destination parameter to prepareProblem(). For instance to prepare
the ROSENBR problem (the one defined by ROSENBR.SIF) and store it in
cache as rbentry, type:
cutermgr.prepareProblem('ROSENBR', destination='rbentry')
Importing the compiled problem interface and its removal from the cache must
now use rbentry instead of ROSENBR:
# Use cutermgr.importProblem()
rb=cutermgr.importProblem('rbentry')
# Remove the compiled problem from cache
cutermgr.clearCache('rbentry')
To check if a problem is in cache under the name rbentry without trying to
import the actual module, use:
if cutermgr.isCached('rbentry'):
...
Specifying problem parameters and sifdecode command line options
Some CUTEr problems have parameters on which the problem itself depends. Often
the dimension of the problem depends on some parameter. Such parameters must
be passed to sifdecode with the -param option. The CUTEr interface handles
such parameters with the sifParams argument to prepareProblem().
Parameters are passed in the form of a Python dictionary, where the key
specifies the name of a parameter. The value of a parameter is converted
using str() to a string and passed to sifdecode’s command line as
-param key=value:
# Prepare the LUBRIFC problem, pass NN=10 to sifdecode
cutermgr.prepareProblem("LUBRIFC", sifParams={'NN': 10})
Arbitrary command line options can be passed to sifdecode by specifying them
in form of a list of strings and passing the list to prepareProblem()
as sifOptions. The following is the equivalent of the last example:
# Prepare the LUBRIFC problem, pass NN=10 to sifdecode
cutermgr.prepareProblem("LUBRIFC", sifOptions=['-param', 'NN=10'])
Specifying variable and constraint ordering
To put nonlinear variables before linear variables set the nvfirst parameter
to True and pass it to func:prepareProblem:
cutermgr.prepareProblem("SOMEPROBLEM", nvfirst=True)
If nvfirst is not specified it defaults to False. In that case no
particular variable ordering is imposed. The variable ordering will be
reflected in the order of variable names returned by the varnames() problem
interface function.
To put equality constraints before inequality constraints set the efirst
parameter to True:
pycutermgr.prepareProblem("SOMEPROBLEM", efirst=True)
Similarly linear constraints can be placed before nonlinear ones by setting
lfirst to True:
pycutermgr.prepareProblem("SOMEPROBLEM", lfirst=True)
Parameters efirst and lfirst default to False meaning that no particular
constraint ordering is imposed. The constraint ordering will be reflected in
the order of constraint names returned by the connames() problem interface
function.
If both efirst and lfirst are set to True, the ordering is a follows:
linear equality constraints followed by linear inequality constraints,
nonlinear equality constraints, and finally nonlinear inequality constraints.
Problem information
The problem information dictionary is returned by the getinfo() problem
interface function. The dictionary has the following entries
name– problem namen– number of variablesm– number of constraints (excluding bounds)x– initial point (1D array of length n)bl– 1D array of length n with lower bounds on variablesbu– 1D array of length n with upper bounds on variablesnnzh– number of nonzero elements in the diagonal and upper triangle of sparse Hessianvartype– 1D integer array of length n storing variable type 0=real, 1=boolean (0 or 1), 2=integernvfirst– boolean flag indicating that nonlinear variables were placed before linear variablessifparams– parameters passed to sifdecode with the sifParams argument toprepareProblem().Noneif no parameters were givensifoptions– additional options passed to sifdecode with the sifOptions argument toprepareProblem().Noneif no additional options were given.
Additional entries are available if the problem has constraints (m>0):
nnzj– number of nonzero elements in sparse Jacobian of constraintsv– 1D array of length m with initial values of Lagrange multiplierscl– 1D array of length m with lower bounds on constraint functionscu– 1D array of length m with upper bounds on constraint functionsequatn– 1D boolean array of length m indicating whether a constraint is an equality constraintlinear– 1D boolean array of length m indicating whether a constraint is a linear constraintefirst– boolean flag indicating that equality constraints were places before inequality constraintslfirst– boolean flag indicating that linear constraints were placed before nonlinear constraints
The names of variables and constraints are returned by the varnames() and
connames() problem interface functions.
Usage statistics
The usage statistics dictionary is returned by the report() problem interface function. The dictionary has the following entries
f– number of objective evaluationsg– number of objective gradient evaluationsH– number of objective Hessian evaluationsHprod– number of Hessian multiplications with a vectortsetup– CPU time used in setuptrun– CPU time used in run
For constrained problems the following additional members are available
c– number of constraint evaluationscg– number of constraint gradient evaluationscH– number of constraint Hessian evaluations
Problem preparation and internal cache organization
The cache ($PYCUTER_CACHE) has one single subdirectory named pycuter holding
all compiled problem interafaces. This way problem interface modules are
accessible as pycuter.NAME because $PYCUTER_CACHE is also listed in
PYTHONPATH.
$PYCUTER_CACHE/pycuter has a dummy __init__.py file generated by
prepareProblem() which specifies that $PYCUTER_CACHE/pycuter
is a Python module. Every problem has its own subdirectory in
$PYCUTER_CACHE/pycuter. In that subdirectory problem decoding (with sifdecode)
and compilation (with gfortran and Python setuptools) take place.
prepareProblem() also generates an __init__.py file for every problem
which takes care of initialization when the problem interface is imported.
The actual binary interaface is in _pycuteritf.so. The __init__.py script
requires the presence of the OUTSDIF.d file where the problem description is
stored. Everything else is neded at compile time only.
Some functions in the _pycuteritf.so module are private (their name starts
with an underscore. These functions are called by wrappers defined in
problem’s __init__.py. An example for this are the sparse CUTEr tools
like scons(). scons() is actually a wrapper defined in __init__.py.
It calls the _scons() function from the problem’s _pycuteritf.so
binary interface module and converts its return values to a
coo_matrix object.
scons() returns the coo_matrix object for J instead
of a NumPy array object. The problem’s _pycuteritf binary module is also
accessible. If the interface module is imported as rb then the _scons()
interface function can be accessed as rb._pycuteritf._scons.
This module does not depend on PyOPUS. It depends only on the
cuteritf module.
-
pyopus.problems.cutermgr.clearCache(cachedName)¶ Removes a cache entry from cache.
Keyword arguments:
- cachedName – cache entry name
-
pyopus.problems.cutermgr.prepareProblem(problemName, destination=None, sifParams=None, sifOptions=None, efirst=False, lfirst=False, nvfirst=False, quiet=True)¶ Prepares a problem interface module, imports and initializes it, and returns a reference to the imported module.
Keyword arguments:
- problemName – CUTEr problem name
- destination – the name under which the compiled problem interface is stored in the cache If not given problemName is used.
- sifParams – parameters passed to sifdecode using the -param command line option
given in the form of a dictionary with parameter name as key. Values
are converted to strings using
str()and every parameter contributes::-param key=str(value)to the sifdecode’s command line options. - sifOptions – additional options passed to sifdecode given in the form of a list of strings.
- efirst – order equation constraints first (default
True) - lfirst – order linear constraints first (default
True) - nvfirst – order nonlinear variables before linear variables (default
False) - quiet – supress output (default
True)
destination must not contain dots because it is a part of a Python module name.
-
pyopus.problems.cutermgr.importProblem(cachedName)¶ Imports and initializes a problem module with CUTEr interface functions. The module must be available in cache (see
prepareProblem()).Keyword arguments:
- cachedName – name under which the problem is stored in cache
-
pyopus.problems.cutermgr.isCached(cachedName)¶ Return
Trueif a problem is in cache.Keyword arguments:
- cachedName – cache entry name
-
pyopus.problems.cutermgr.updateClassifications(verbose=False)¶ Updates the list of problem classifications from SIF files. Collects the CUTEr problem classification strings.
- verbose – if set to
True, prints output as files are scanned
- Every SIF file contains a line of the form
-something- classification -code-- Code has the following format
OCRr-GI-N-M
O (single letter) - type of objective
N.. no objective function definedC.. constant objective functionL.. linear objective functionQ.. quadratic objective functionS.. objective function is a sum of squaresO.. none of the above
C (single letter) - type of constraints
U.. unconstrainedX.. equality constraints on variablesB.. bounds on variablesN.. constraints represent the adjacency matrix of a (linear) networkL.. linear constraintsQ.. quadratic constraintsO.. more general than any of the above
R (single letter) - problem regularity
R.. regular - first and second derivatives exist and are continuousI.. irregular problem
r (integer) - degree of the highest derivatives provided analytically within the problem description, can be 0, 1, or 2
G (single letter) - origin of the problem
A.. academic (created for testing algorithms)M.. modelling exercise (actual value not used in practical application)R.. real-world problem
I (single letter) - problem contains explicit internal variables
Y.. yesN.. no
N (integer or
V) - number of variables,V= can be set by userM (integer or
V) - number of constraints,V= can be set by user- verbose – if set to
-
pyopus.problems.cutermgr.problemProperties(problemName)¶ Returns problem properties (uses the CUTEr problem classification string).
problemName – problem name
Returns a dictionary with the following members:
objective– objective type codeconstraints– constraints type coderegular–Trueif problem is regulardegree– highest degree of analytically available derivativeorigin– problem origin codeinternal–Trueif problem has internal variablesn– number of variables (None= can be set by the user)m– number of constraints (None= can be set by the user)
-
pyopus.problems.cutermgr.findProblems(objective=None, constraints=None, regular=None, degree=None, origin=None, internal=None, n=None, userN=None, m=None, userM=None)¶ Returns the problem names of problems that match the given requirements. The search is based on the CUTEr problem classification string.
- objective – a string containg one or more letters (NCLQSO) specifying the type of the objective function
- constraints – a string containg one or more letters (UXBNLQO) the type of the constraints
- regular – a boolean,
Trueif the problem must be regular,Falseif it must be irregular - degree – list of the form [min, max] specifying the minimum and the maximum number of analytically available derivatives
- origin – a string containg one or more letters (AMR) specifying the origin of the problem
- internal – a boolean,
Trueif the problem must have internal variables,Falseif internal variables are not allowed - n – a list of the form [min, max] specifying the lowest and the highest allowed number of variables
- userN –
Trueof the problems must have user settable number of variables,Falseif the number must be hardcoded - m – a list of the form [min, max] specifying the lowest and the highest allowed number of constraints
- userM –
Trueof the problems must have user settable number of variables,Falseif the number must be hardcoded
Problems with a user-settable number of variables/constraints match any given n / m.
Returns the problem names of problems that matched the given requirements.
If a requirement is not given, it is not applied.
See
updateClassifications()for details on the letters used in the requirements.
Unconstrained problem example file cutermgru.py in folder demo/problems/
# Demonstration of CUTEr use - unconstrained problem (ROSENBR)
from pyopus.problems import cutermgr
from numpy import array
if __name__ == '__main__':
# Clear cache - this is not neccessary because the cache entry is removed
# by prepareProblem() before the problem interface is built.
cutermgr.clearCache('ROSENBR')
# Prepare two problems
cutermgr.prepareProblem('ROSENBR')
# These two are equivalent and demonstrate the use of sifParams and sifOptions.
# prepareProblem("LUBRIFC", sifParams={'NN': 10})
# prepareProblem("LUBRIFC", sifOptions=['-param', 'NN=10'])
# Import ROSENBR (unconstrained problem)
# prepareProblem() returns a reference to the imported module.
# Use this only if the problem interface is already available in the cache.
uproblem=cutermgr.importProblem('ROSENBR')
# ROSENBR
#
# f = (1 - x1)^2 + 100 (x2 - x1^2)^2
#
# g = [ -2 (1 - x1) - 400 (x2 - x1^2) x1 200 (x2 - x1^2) ]
#
# [ 2 - 400 (x2 - x1^2) + 800 x1^2 -400 x1 ]
# H = [ ]
# [ -400 x1 200 ]
#
#
# at x0 = [ -1.2 1.0 ]
#
# f = 24.2
# g = [ -215.6 -88.0 ]
#
# [ 1330.0 480.0 ]
# H = [ ]
# [ 480.0 200.0 ]
print("Unconstrained problem demo")
info=uproblem.getinfo()
print("Problem name : ", info['name'])
print("Problem size : ", info['n'])
print("Constraint count : ", info['m'])
print("NNZ in UT Hessian : ", info['nnzh'])
print("Lower bounds : ", info['bl'])
print("Upper bounds : ", info['bu'])
print("Initial point : ", info['x'])
print("Variable type : ", info['vartype'])
print("Variable names : ", uproblem.varnames())
print("Sifdecode params : ", info['sifparams'])
print("Sifdecode options : ", info['sifoptions'])
print("Nonl. vars. first : ", info['nvfirst'])
x0=info['x']
print("\nEvaluating objective at x0")
f=uproblem.obj(x0)
print("f(x0)=", f)
print("\nEvaluating function and gradient")
(f, g)=uproblem.obj(x0, True)
print("f(x0)=", f)
print("g(x0)=", g)
print("\nEvaluating function and constraints")
(f, c)=uproblem.objcons(x0)
print("f(x0)=", f)
print("c(x0)=", c)
print("\nEvaluating constraints")
c=uproblem.cons(x0)
print("c(x0)=", c)
print("\nEvaluating constraints and Jacobian")
(c, J)=uproblem.cons(x0, True)
print("c(x0)=", c)
print("J(x0)=", J)
print("\nEvaluating function gradient and constraints Jacobian")
(g, J)=uproblem.lagjac(x0)
print("g(x0)=", g)
print("J(x0)=", J)
print("\nEvaluating Hessian of objective for unconstrained problem")
H=uproblem.hess(x0)
print("H(x0)=", H)
print("\nEvaluating Hessian of objective")
H=uproblem.ihess(x0)
print("H(x0)=", H)
print("\nEvaluating Hessian at x0 times [2.0, 2.0]")
r=uproblem.hprod(array([2.0, 2.0]), x0)
print("H(x0)*[2.0, 2.0]=", r)
print("\nEvaluating previous Hessian times [2.0, 2.0]")
r=uproblem.hprod(array([2.0, 2.0]))
print("H*[2.0, 2.0]=", r)
print("\nEvaluating gradient of objective at x0 and Hessian")
(g, H)=uproblem.gradhess(x0)
print("g(x0)=", g)
print("H(x0)=", H)
# Sparse Jacobian cannot be obtained for unconstrained problems because sparse
# matrices of size 0-by-n are not supported in SciPy.
print("\nEvaluating sparse Hessian of objective for unconstrained problem")
H=uproblem.sphess(x0)
print("H(x0)=", H.todense())
print("\nEvaluating sparse Hessian of objective")
H=uproblem.isphess(x0)
print("H(x0)=", H.todense())
print("\nEvaluating gradient and sparse Hessian of objective")
(g, J)=uproblem.gradsphess(x0)
print("g(x0)=", g)
print("J(x0)=", J.todense())
print("\nCollecting report")
rep=uproblem.report()
print("Setup time : ", rep['tsetup'])
print("Run time : ", rep['trun'])
print("Num. of f eval : ", rep['f'])
print("Num. of g eval : ", rep['g'])
print("Num. of H eval : ", rep['H'])
print("Num. of H prod : ", rep['Hprod'])
Constrained problem example file cutermgrc.py in folder demo/problems/
# Demonstration of PyCUTEr use - constrained problem (HS71)
from pyopus.problems import cutermgr
from numpy import array
if __name__ == '__main__':
# Clear cache - this is not neccessary because the cache entry is removed
# by prepareProblem() before the problem interface is built.
cutermgr.clearCache('HS71')
# Prepare two problems
cutermgr.prepareProblem('HS71')
# Import HS71 (unconstrained problem)
# prepareProblem() returns a reference to the imported module.
# Use this only if the problem interface is already available in the cache.
cproblem=cutermgr.importProblem('HS71')
# HS71
#
# f = x1 x4 (x1 + x2 + x3) + x3
# subject to
# (C1) x1 x2 x3 x4 - 25 >= 0
# (C2) x1^2 + x2^2 + x3^2 + x4^2 - 40 = 0
# (bounds) 1 <= x1, x2, x3, x4 <= 5
#
# g = [ x4 (x1 + x2 + x3) + x1 x4 x1 x4 x1 x4 + 1 x1 (x1 + x2 + x3) ]
#
# [ 2 x4 x4 x4 2 x1 + x2 + x3 ]
# [ ]
# [ x4 0 0 x1 ]
# H = [ ]
# [ x4 0 0 x1 ]
# [ ]
# [ 2 x1 + x2 + x3 x1 x1 0 ]
#
# [ x2 x3 x4 x1 x3 x4 x1 x2 x4 x1 x2 x3 ]
# J = [ ]
# [ 2*x1 2 x2 2 x3 2 x4 ]
#
# [ 0 x3 x4 x2 x4 x2 x3 ]
# [ ]
# [ x3 x4 0 x1 x4 x1 x3 ]
# HC1 = [ ]
# [ x2 x4 x1 x4 0 x1 x2 ]
# [ ]
# [ x2 x3 x1 x3 x1 x2 0 ]
#
# [ 2 0 0 0 ]
# [ ]
# [ 0 2 0 0 ]
# HC2 = [ ]
# [ 0 0 2 0 ]
# [ ]
# [ 0 0 0 2 ]
#
# at x0 = [ 1.0 5.0 5.0 1.0 ]
#
# f = 16.0
# c1 = 0.0
# c2 = 12.0
#
# g = [ 12.0 1.0 2.0 11.0 ]
#
# [ 2.0 1.0 1.0 12.0 ]
# [ ]
# [ 1.0 0.0 0.0 1.0 ]
# H = [ ]
# [ 1.0 0.0 0.0 1.0 ]
# [ ]
# [ 12.0 1.0 1.0 0.0 ]
#
# [ 25.0 5.0 5.0 25.0 ]
# J = [ ]
# [ 2.0 10.0 10.0 2.0 ]
#
# [ 0.0 5.0 5.0 25.0 ]
# [ ]
# [ 5.0 0.0 1.0 5.0 ]
# HC1 = [ ]
# [ 5.0 1.0 0.0 5.0 ]
# [ ]
# [ 25.0 5.0 5.0 0.0 ]
print("Constrained problem demo")
info=cproblem.getinfo()
print("Problem name : ", info['name'])
print("Problem size : ", info['n'])
print("Constraint count : ", info['m'])
print("NNZ in UT Hessian : ", info['nnzh'])
print("Lower bounds : ", info['bl'])
print("Upper bounds : ", info['bu'])
print("Initial point : ", info['x'])
print("Variable type : ", info['vartype'])
print("Variable names : ", cproblem.varnames())
print("Sifdecode params : ", info['sifparams'])
print("Sifdecode options : ", info['sifoptions'])
print("Constraint names : ", cproblem.connames())
print("Eq. constr. first : ", info['efirst'])
print("Lin. constr. first : ", info['lfirst'])
print("NNZ in Jacobian : ", info['nnzj'])
print("Equality constr. : ", info['equatn'])
print("Linear constr. : ", info['linear'])
print("Lower constraint : ", info['cl'])
print("Upper constraint : ", info['cu'])
print("Init. Lagr. mult. : ", info['v'])
x0=info['x']
print("\nEvaluating objective at x0")
f=cproblem.obj(x0)
print("f(x0)=", f)
print("\nEvaluating function and gradient")
(f, g)=cproblem.obj(x0, True)
print("f(x0)=", f)
print("g(x0)=", g)
print("\nEvaluating function and constraints")
(f, c)=cproblem.objcons(x0)
print("f(x0)=", f)
print("c(x0)=", c)
print("\nEvaluating constraints")
c=cproblem.cons(x0)
print("c(x0)=", c)
print("\nEvaluating single constraint (0)")
c0=cproblem.cons(x0, False, 0)
print("c0(x0) : ", c0)
print("\nEvaluating constraints and Jacobian")
(c, J)=cproblem.cons(x0, True)
print("c(x0)=", c)
print("J(x0)=", J)
print("\nEvaluating single constraint (0) and its gradient")
(c0, gc0)=cproblem.cons(x0, True, 0)
print("c0(x0)=", c0)
print("gc0(x0)=", gc0)
print("\nEvaluating function gradient and constraints Jacobian")
(g, J)=cproblem.lagjac(x0)
print("g(x0)=", g)
print("J(x0)=", J)
print("\nEvaluating gradient of Lagrangian at v=[1, 1] and constraints Jacobian")
(g, J)=cproblem.lagjac(x0, array([1.0, 1.0]))
print("gl(x0,[1.0, 1.0])=", g)
print("J(x0)=", J)
print("\nEvaluating product of previous J and [1, 1, 1, 1]")
r=cproblem.jprod(False, array([1.0, 1.0, 1.0, 1.0]))
print("J*[1.0, 1.0, 1.0, 1.0]=", r)
print("\nEvaluating product of J at x0 and [1, 1, 1, 1]")
r=cproblem.jprod(False, array([1.0, 1.0, 1.0, 1.0]), x0)
print("J(x0)*[1.0, 1.0, 1.0, 1.0]=", r)
print("\nEvaluating product of previous transposed J and [1, 1]")
r=cproblem.jprod(True, array([1.0, 1.0]))
print("JT*[1.0, 1.0]=", r)
print("\nEvaluating product of transposed J at x0 and [1, 1]")
r=cproblem.jprod(True, array([1.0, 1.0]), x0)
print("JT(x0)*[1.0, 1.0]=", r)
print("\nEvaluating Hessian of Lagrangian at [1, 1] for constrained problem")
Hl=cproblem.hess(x0, array([1.0, 1.0]))
print("Hl(x0, [1.0, 1.0])-", Hl)
print("\nEvaluating Hessian of objective")
H=cproblem.ihess(x0)
print("H(x0)=", H)
print("\nEvaluating Hessian of constraint (0)")
H0=cproblem.ihess(x0, 0)
print("H0(x0)=", H0)
print("\nEvaluating Hessian of constraint (1)")
H1=cproblem.ihess(x0, 1)
print("H1(x0)=", H1)
print("\nEvaluating Hessian of the Lagrangian at (x0, [1.0, 1.0]) times [2.0, 2.0, 2.0, 2.0]")
r=cproblem.hprod(array([2.0, 2.0, 2.0, 2.0]), x0, array([1.0, 1.0]))
print("Hl(x0, [1.0, 1.0])*[2.0, 2.0, 2.0, 2.0]=", r)
print("\nEvaluating previous Hess. of the Lagr. times [2.0, 2.0, 2.0, 2.0]")
r=cproblem.hprod(array([2.0, 2.0, 2.0, 2.0]))
print("Hl*[2.0, 2.0, 2.0, 2.0]=", r)
print("\nEvaluating gradient of Lagrangian at (x0, [1.0, 1.0]), Jacobian, and Hessian of Lagrangian")
(gl, J, Hl)=cproblem.gradhess(x0, array([1.0, 1.0]), True)
print("gl(x0, [1.0, 1.0])=", gl)
print("J(x0)=", J)
print("Hl(x0, [1.0, 1.0])=", Hl)
print("\nEvaluating grad. of obj. at x0, Jacobian, and Hessian of Lagrangian")
(g, J, Hl)=cproblem.gradhess(x0, array([1.0, 1.0]), False)
print("g(x0)=", g)
print("J(x0)=", J)
print("Hl(x0)=", Hl)
print("\nEvaluating constraints and sparse Jacobian")
(c, J)=cproblem.scons(x0)
print("c(x0)=", c)
print("J(x0)=", J.todense())
print("\nEvaluating constraint (0) and its sparse gradient")
(c0, gc0)=cproblem.scons(x0, 0)
print("c0(x0)=", c0)
print("gc0(x0)=", gc0.todense())
print("\nEvaluating sparse objective gradient and sparse Jacobian")
(g, J)=cproblem.slagjac(x0)
print("g(x0)=", g.todense())
print("J(x0)=", J.todense())
print("\nEvaluating sparse Lagrangian gradient and sparse Jacobian at (x0, [1.0, 1.0])")
(gl, J)=cproblem.slagjac(x0, array([1.0, 01.0]))
print("gl(x0)=", gl.todense())
print("J(x0)=", J.todense())
print("\nEvaluating sparse Hessian of Lagrangian at (x0, [1, 1]) for constrained problem")
Hl=cproblem.sphess(x0, array([1.0, 1.0]))
print("Hl(x0, [1.0, 1.0])=", Hl.todense())
print("\nEvaluating sparse Hessian of objective")
H=cproblem.isphess(x0)
print("H(x0)=", H.todense())
print("\nEvaluating sparse Hessian of single constraint (0)")
H0=cproblem.isphess(x0, 0)
print("H0(x0)=", H0.todense())
print("\nEvaluating sparse Hessian of single constraint (1)")
H1=cproblem.isphess(x0, 1)
print("H0(x0)=", H1.todense())
print("\nEvaluating grad. of obj., constr. Jac., and Hess. of Lagr. at (x0, [0.0, 0.0])")
(g, J, Hl)=cproblem.gradsphess(x0, array([1.0, 1.0]))
print("g(x0)=", g.todense())
print("J(x0)=", J.todense())
print("Hl(x0, [1.0, 1.0])=", Hl.todense())
print("\nEvaluating grad. of Lagr., constr. Jac., and Hess. of Lagr. at (x0, [0.0, 0.0])")
(gl, J, Hl)=cproblem.gradsphess(x0, array([1.0, 1.0]), True)
print("gl(x0)=", gl.todense())
print("J(x0)=", J.todense())
print("Hl(x0, [1.0, 1.0])=", Hl.todense())
print("\nCollecting report")
rep=cproblem.report()
print("Setup time : ", rep['tsetup'])
print("Run time : ", rep['trun'])
print("Num. of f eval : ", rep['f'])
print("Num. of g eval : ", rep['g'])
print("Num. of H eval : ", rep['H'])
print("Num. of H prod : ", rep['Hprod'])
print("Num. of c eval : ", rep['c'])
print("Num. of cg eval : ", rep['cg'])
print("Num. of cH eval : ", rep['cH'])